

For example, does a slight change in FA threshold dramatically change the appearance and connectivity profile of a certain fiber bundle? Another disadvantage of pre-computing streamline datasets is that parameters used for tractography may not be the same across different subjects and different regions of the same brain ( Pierpaoli et al., 1996). However, since calculation and display steps are not linked, a major disadvantage of this approach is that the user cannot directly observe the tracking process.
#Medinria vide software
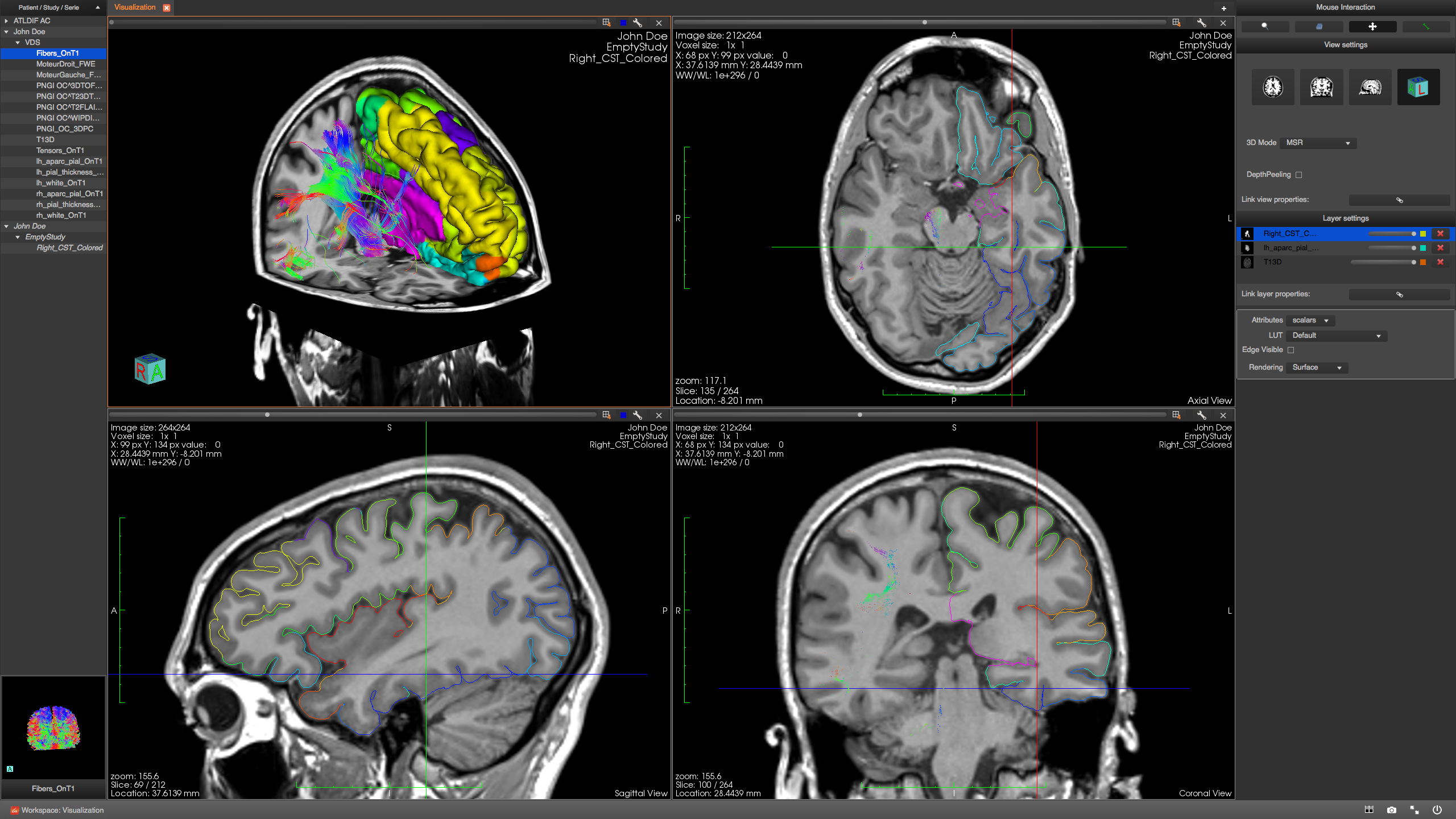
The most common approach is to first reconstruct streamlines using a fixed set of parameters and then visualize the results using one of the different software packages (e.g., MRtrix ExploreDTI TrackVis Camino DTI-Studio MedINRIA, MITK BrainVISA amongst others). Typically, streamline generation and streamline visualization are done separately. These techniques fall into the streamline fiber tracking family of techniques ( Mori et al., 1999 Tournier et al., 2011). The classical way to perform such reconstruction is by following the main diffusion tensor (DT) or orientation distribution function (ODF) direction at each voxel. One of the main features of diffusion MRI consists of the reconstruction of the white matter (WM) fiber pathways in the brain using a computerized technique called tractography. Finally, we show how our RTT tool facilitates neurosurgical planning and allows one to find fibers that infiltrate tumor areas, otherwise missing when using the standard default tracking parameters.
#Medinria vide Offline
We qualitatively illustrate and quantitatively evaluate our novel multi-peak RTT technique on phantom and human datasets in comparison with the state of the art offline tractography from MRtrix, which is robust to fiber crossings. The technique runs on a single Computer Processing Unit (CPU) without the need for Graphical Unit Processing (GPU) programming. To achieve such real-time performance, we propose a novel evolution equation based on the upsampled principal directions, also called peaks, extracted at each voxel of the dMRI dataset. In this work, we propose a real-time fiber tracking (RTT) tool which can instantaneously compute and display streamlines.

Having access to tractography parameters can thus be advantageous, as it will help in better isolating those which are sensitive to certain streamline features and potentially converge on optimal settings which are area-specific. As a result, a slight change in tracking parameters may return different connectivity profiles and complicate the interpretation of the results. tumor patient), optimal tracking parameters can be dramatically different. Therefore, depending on the area of interest or subject (e.g., healthy control vs. However, the scale and curvature of fiber bundles can vary from region to region in the brain. Most publicly available tractography techniques and most studies are based on a fixed set of tractography parameters. Tractography is nowadays central in structural connectivity since it is the only non-invasive technique to obtain information about brain wiring. The computerized process of reconstructing white matter tracts from diffusion MRI (dMRI) data is often referred to as tractography. 5Division of Neurosurgery and Neuro-Oncology, Faculty of Medicine and Health Science, University of Sherbrooke, Sherbrooke, QC, Canada.4Department of Diagnostic Radiology, University of Sherbrooke, Sherbrooke, QC, Canada.3Sherbrooke Connectivity Imaging Lab, Computer Science Department, Faculty of Science, University of Sherbrooke, Sherbrooke, QC, Canada.2Department of Nuclear Medecine and Radiobiology, University of Sherbrooke, Sherbrooke, QC, Canada.
1Centre de Recherche CHUS, University of Sherbrooke, Sherbrooke, QC, Canada.Maxime Chamberland 1,2,3 *, Kevin Whittingstall 1,2,4, David Fortin 1,5, David Mathieu 5 and Maxime Descoteaux 1,3 *


 0 kommentar(er)
0 kommentar(er)
